Design of Experiment: Optimize perovskite solar cells efficiency (TURBO)
This notebook is made to use optimPV for experimental design. Here, we show how to load some data from a presampling, and how to use optimPV to suggest the next set of experiment using Bayesian optimization. The goal here is to optimize the processing conditions for a perovskite solar cell to maximize the power conversion efficiency (PCE).
Note: The data used here is real data generated in the i-MEET and HI-ERN labs at the university of Erlangen-Nuremberg (FAU) by Jiyun Zhang for the paper: Autonomous Optimization of Air-Processed Perovskite Solar Cell in a 6D Parameter Space
[1]:
# Import necessary libraries
import warnings, os, sys, torch
# remove warnings from the output
os.environ["PYTHONWARNINGS"] = "ignore"
warnings.filterwarnings(action='ignore', category=FutureWarning)
warnings.filterwarnings(action='ignore', category=UserWarning)
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import ax
from ax.utils.notebook.plotting import init_notebook_plotting
init_notebook_plotting() # for Jupyter notebooks
try:
from optimpv import *
from optimpv.optimizers.axBOtorch.axUtils import *
except Exception as e:
sys.path.append('../') # add the path to the optimpv module
from optimpv import *
from optimpv.optimizers.axBOtorch.axUtils import *
[INFO 01-20 09:42:10] ax.utils.notebook.plotting: Injecting Plotly library into cell. Do not overwrite or delete cell.
[INFO 01-20 09:42:10] ax.utils.notebook.plotting: Please see
(https://ax.dev/tutorials/visualizations.html#Fix-for-plots-that-are-not-rendering)
if visualizations are not rendering.
Get the data
[2]:
# Define the path to the data
data_dir =os.path.join(os.path.abspath('../'),'Data','6D_pero_opti') # path to the data directory
# Load the data
df = pd.read_csv(os.path.join(data_dir,'6D_pero_opti.csv'),sep=r'\s+') # load the data
# Display some information about the data
print(df.describe())
Spin_Speed_1 Duration_t1 Spin_Speed_2 Dispense_Speed Duration_t3 \
count 76.000000 76.000000 76.000000 76.000000 76.000000
mean 1520.486842 18.276316 2273.289474 234.697368 17.315789
std 458.510240 7.000188 501.488845 83.133470 7.919950
min 540.000000 5.000000 1021.000000 16.000000 5.000000
25% 1203.500000 13.750000 2019.500000 201.750000 10.750000
50% 1511.000000 18.000000 2385.500000 247.500000 17.000000
75% 1801.250000 22.000000 2671.000000 277.500000 23.250000
max 2579.000000 34.000000 3000.000000 396.000000 35.000000
Spin_Speed_3 Jsc Voc FF Pmax Vmpp \
count 76.000000 76.000000 76.000000 76.000000 76.000000 76.000000
mean 3717.789474 24.391395 1.021157 0.724692 18.320711 0.825778
std 917.207346 1.872929 0.108197 0.055290 3.486546 0.105413
min 2000.000000 12.231000 0.616300 0.448800 4.690000 0.463000
25% 3002.000000 24.173500 0.987650 0.704400 17.477000 0.785750
50% 3825.000000 24.615000 1.038750 0.734700 18.851500 0.835000
75% 4471.500000 25.279500 1.100725 0.759925 20.347250 0.895000
max 5000.000000 25.938000 1.162200 0.810900 23.729000 0.990100
Rseries Rshunt
count 76.000000 76.000000
mean 3720.112882 2763.793289
std 1487.516713 1956.263177
min 92.129000 2.990000
25% 3146.452500 228.750000
50% 3739.157000 3290.000000
75% 4567.406250 4285.000000
max 6513.151000 6860.000000
Define the parameters for the simulation
[3]:
params = [] # list of parameters to be optimized
Spin_Speed_1 = FitParam(name = 'Spin_Speed_1', value = 1000, bounds = [500,3000], value_type = 'int', display_name='Spin Speed 1', unit='rpm', axis_type = 'linear')
params.append(Spin_Speed_1)
Duration_t1 = FitParam(name = 'Duration_t1', value = 10, bounds = [5,35], value_type = 'int', display_name='Duration t1', unit='s', axis_type = 'linear')
params.append(Duration_t1)
Spin_Speed_2 = FitParam(name = 'Spin_Speed_2', value = 1000, bounds = [1000,3000], value_type = 'int', display_name='Spin Speed 2', unit='rpm', axis_type = 'linear')
params.append(Spin_Speed_2)
Dispense_Speed = FitParam(name = 'Dispense_Speed', value = 100, bounds = [10,400], value_type = 'int', display_name='Dispense Speed', unit='rpm', axis_type = 'linear')
params.append(Dispense_Speed)
Duration_t3 = FitParam(name = 'Duration_t3', value = 10, bounds = [5,35], value_type = 'int', display_name='Duration t3', unit='s', axis_type = 'linear')
params.append(Duration_t3)
Spin_Speed_3 = FitParam(name = 'Spin_Speed_3', value = 3000, bounds = [2000,5000], value_type = 'int', display_name='Spin Speed 3', unit='rpm', axis_type = 'linear')
params.append(Spin_Speed_3)
Run the optimization
[4]:
# Define the Agent and the target metric/loss function
from optimpv.general.SuggestOnlyAgent import SuggestOnlyAgent
suggest = SuggestOnlyAgent(params,exp_format='Pmax',minimize=False,tracking_exp_format=['Jsc','Voc','FF'],name=None)
[5]:
from optimpv.optimizers.axBOtorch.axBOtorchOptimizer import axBOtorchOptimizer
from botorch.acquisition.logei import qLogNoisyExpectedImprovement
from ax.adapter.transforms.standardize_y import StandardizeY
from ax.adapter.transforms.unit_x import UnitX
from ax.adapter.transforms.remove_fixed import RemoveFixed
from ax.adapter.transforms.log import Log
from ax.generators.torch.botorch_modular.utils import ModelConfig
from ax.generators.torch.botorch_modular.surrogate import SurrogateSpec
from gpytorch.kernels import MaternKernel
from gpytorch.kernels import ScaleKernel
from botorch.models.fully_bayesian import SaasFullyBayesianSingleTaskGP
model_gen_kwargs_list = None
parameter_constraints = None
model_kwargs_list = [{"torch_device":torch.device("cuda" if torch.cuda.is_available() else "cpu"),'botorch_acqf_class':qLogNoisyExpectedImprovement,'transforms':[RemoveFixed, Log, UnitX, StandardizeY],'surrogate_spec':SurrogateSpec(model_configs=[ModelConfig(botorch_model_class=SaasFullyBayesianSingleTaskGP,covar_module_class=ScaleKernel, covar_module_options={'base_kernel':MaternKernel(nu=2.5, ard_num_dims=len(params))})])}]
model_kwargs_list =None
# Define the optimizer
optimizer = axBOtorchOptimizer(params = params, agents = suggest, models = ['BOTORCH_MODULAR'],n_batches = [1], batch_size = [6], ax_client = None, max_parallelism = -1, model_kwargs_list = model_kwargs_list, model_gen_kwargs_list = None, name = 'ax_opti',suggest_only = True,existing_data=df,verbose_logging=True)
[6]:
# Run the optimization
best_value_previous_step = 22.973
kwargs_turbo_state = {'length': 0.4, 'success_counter': 1, 'failure_counter': 1 }
turbo_state_params = optimizer.optimize_turbo(kwargs_turbo={"best_value": best_value_previous_step}, kwargs_turbo_state=kwargs_turbo_state) # run the optimization with turbo
# The current state of the turbo optimization is stored in the kwargs_turbo_state parameter, which can be used to continue the optimization in the next step
[INFO 01-20 09:42:11] optimpv.axBOtorchOptimizer: Starting optimization with 1 batches and a total of 6 trials
[INFO 01-20 09:42:11] optimpv.axBOtorchOptimizer: Using existing data for initialization
[INFO 01-20 09:42:11] optimpv.axBOtorchOptimizer: Suggesting 6 trials without running the agents.
[WARNING 01-20 09:42:12] ax.api.client: Metric IMetric('Jsc') not found in optimization config, added as tracking metric.
[WARNING 01-20 09:42:12] ax.api.client: Metric IMetric('Voc') not found in optimization config, added as tracking metric.
[WARNING 01-20 09:42:12] ax.api.client: Metric IMetric('FF') not found in optimization config, added as tracking metric.
[INFO 01-20 09:42:12] optimpv.axBOtorchOptimizer: Suggesting 6 trials without running the agents
[7]:
# get the best parameters and update the params list in the optimizer and the agent
ax_client = optimizer.ax_client # get the ax client
optimizer.update_params_with_best_balance() # update the params list in the optimizer with the best parameters
suggest.params = optimizer.params # update the params list in the agent with the best parameters
print("Best parameters found:")
for p in optimizer.params:
print(f"{p.name}: {p.value} {p.unit} ")
Best parameters found:
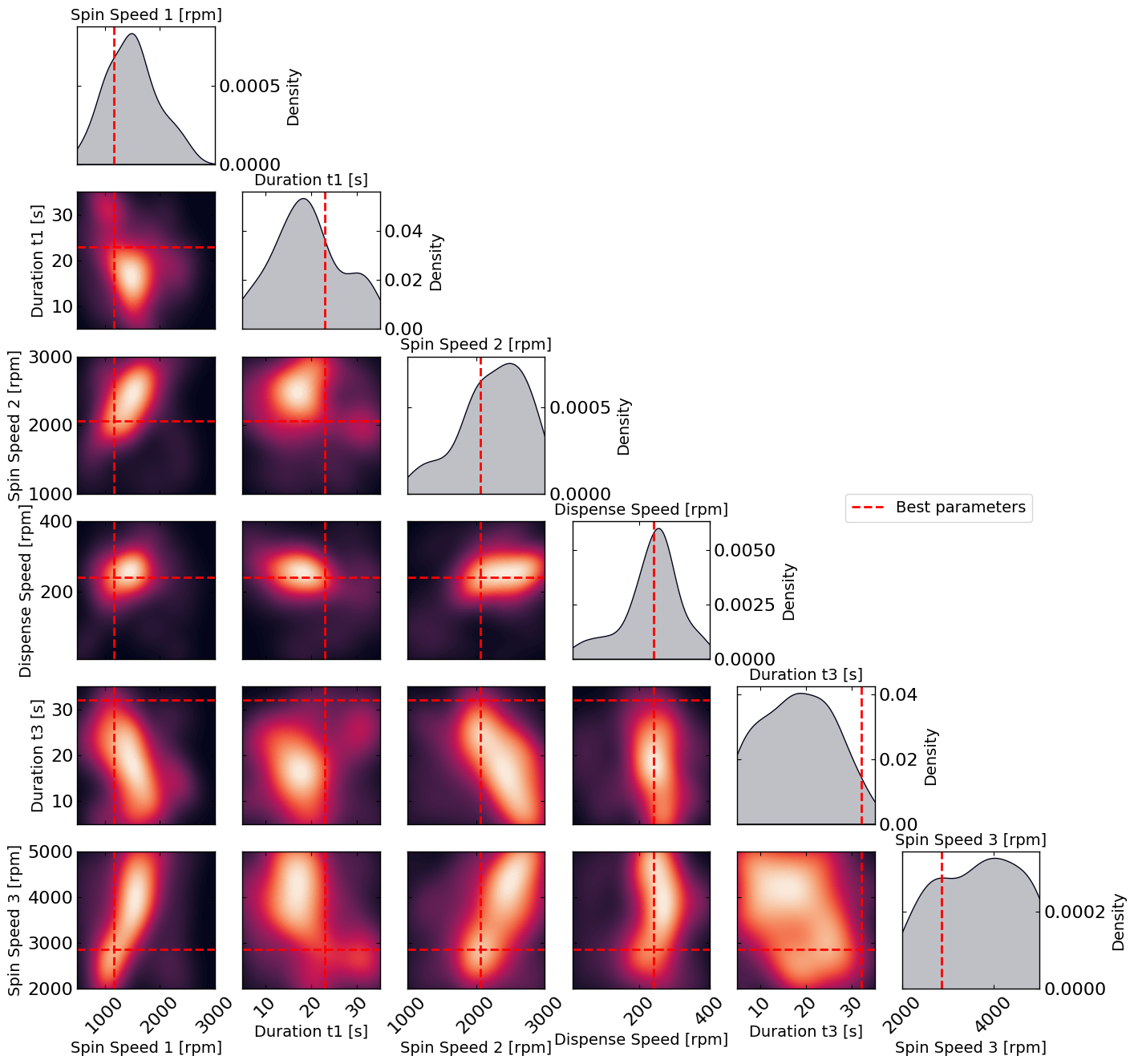
Spin_Speed_1: 1165 rpm
Duration_t1: 23 s
Spin_Speed_2: 2063 rpm
Dispense_Speed: 241 rpm
Duration_t3: 32 s
Spin_Speed_3: 2863 rpm
[8]:
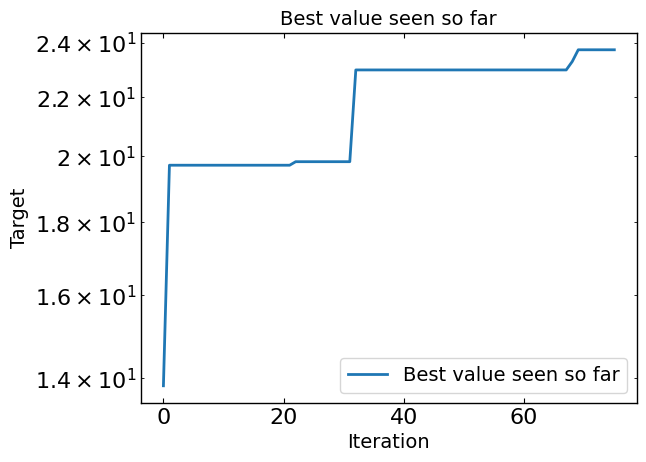
# Plot optimization results
data = ax_client.summarize()
all_metrics = optimizer.all_metrics
plt.figure()
plt.plot(np.maximum.accumulate(data[all_metrics]), label="Best value seen so far")
plt.yscale("log")
plt.xlabel("Iteration")
plt.ylabel("Target")
plt.legend()
plt.title("Best value seen so far")
print("Best value seen so far is ", max(data[all_metrics[0]]), "at iteration ", int(data[all_metrics[0]].idxmin()))
plt.show()
Best value seen so far is 23.729 at iteration 27
[9]:
# print the parameters to that are running in data
to_run_next = data[data['trial_status'] == 'RUNNING']
print("Parameters to run next:")
print(to_run_next)
Parameters to run next:
trial_index arm_name trial_status Jsc Voc FF Pmax Spin_Speed_1 \
76 76 76_0 RUNNING NaN NaN NaN NaN 1154
77 77 77_0 RUNNING NaN NaN NaN NaN 1146
78 78 78_0 RUNNING NaN NaN NaN NaN 1148
79 79 79_0 RUNNING NaN NaN NaN NaN 1181
80 80 80_0 RUNNING NaN NaN NaN NaN 1165
81 81 81_0 RUNNING NaN NaN NaN NaN 1164
Duration_t1 Spin_Speed_2 Dispense_Speed Duration_t3 Spin_Speed_3
76 29 1883 262 31 2649
77 25 1924 256 32 2734
78 31 1972 242 33 2633
79 31 1958 263 32 2777
80 23 1976 253 33 2692
81 23 1962 268 34 2712
[10]:
cards = ax_client.compute_analyses(display=True)
This analysis provides an overview of the entire optimization process. It includes visualizations of the results obtained so far, insights into the parameter and metric relationships learned by the Ax model, diagnostics such as model fit, and health checks to assess the overall health of the experiment.
Result Analyses provide a high-level overview of the results of the optimization process so far with respect to the metrics specified in experiment design.
These pair of plots visualize the metric effects for each arm, with the Ax model predictions on the left and the raw observed data on the right. The predicted effects apply shrinkage for noise and adjust for non-stationarity in the data, so they are more representative of the reproducible effects that will manifest in a long-term validation experiment.
Modeled Arm Effects on Pmax
Modeled effects on Pmax. This plot visualizes predictions of the true metric changes for each arm based on Ax's model. This is the expected delta you would expect if you (re-)ran that arm. This plot helps in anticipating the outcomes and performance of arms based on the model's predictions. Note, flat predictions across arms indicate that the model predicts that there is no effect, meaning if you were to re-run the experiment, the delta you would see would be small and fall within the confidence interval indicated in the plot.
Observed Arm Effects on Pmax
Observed effects on Pmax. This plot visualizes the effects from previously-run arms on a specific metric, providing insights into their performance. This plot allows one to compare and contrast the effectiveness of different arms, highlighting which configurations have yielded the most favorable outcomes.
Summary for ax_opti
High-level summary of the `Trial`-s in this `Experiment`
| trial_index | arm_name | trial_status | Jsc | Voc | FF | Pmax | Spin_Speed_1 | Duration_t1 | Spin_Speed_2 | Dispense_Speed | Duration_t3 | Spin_Speed_3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0_0 | COMPLETED | 24.507 | 0.9100 | 0.6201 | 13.830 | 2404 | 15 | 1745 | 203 | 13 | 4494 |
| 1 | 1 | 1_0 | COMPLETED | 25.733 | 1.0090 | 0.7592 | 19.712 | 1835 | 17 | 2289 | 289 | 11 | 4389 |
| 2 | 2 | 2_0 | COMPLETED | 24.692 | 1.0321 | 0.7376 | 18.797 | 1067 | 33 | 2352 | 234 | 5 | 2861 |
| 3 | 3 | 3_0 | COMPLETED | 24.195 | 0.8197 | 0.6506 | 12.903 | 2276 | 18 | 1821 | 26 | 15 | 3241 |
| 4 | 4 | 4_0 | COMPLETED | 24.268 | 0.9951 | 0.7684 | 18.556 | 2147 | 30 | 2816 | 290 | 29 | 4981 |
| 5 | 5 | 5_0 | COMPLETED | 24.481 | 1.0100 | 0.7264 | 17.962 | 1152 | 28 | 2734 | 121 | 8 | 4856 |
| 6 | 6 | 6_0 | COMPLETED | 25.575 | 0.9926 | 0.7377 | 18.726 | 1240 | 23 | 2993 | 190 | 19 | 2560 |
| 7 | 7 | 7_0 | COMPLETED | 23.995 | 0.6850 | 0.6336 | 10.414 | 2471 | 21 | 1394 | 138 | 29 | 2046 |
| 8 | 8 | 8_0 | COMPLETED | 24.283 | 0.9940 | 0.7185 | 17.342 | 907 | 9 | 2766 | 314 | 22 | 4323 |
| 9 | 9 | 9_0 | COMPLETED | 12.231 | 0.8321 | 0.5819 | 5.922 | 2034 | 18 | 1021 | 90 | 7 | 4776 |
| 10 | 10 | 10_0 | COMPLETED | 23.558 | 0.9297 | 0.6919 | 15.154 | 540 | 19 | 2383 | 16 | 23 | 4632 |
| 11 | 11 | 11_0 | COMPLETED | 25.214 | 0.9921 | 0.7637 | 19.105 | 1532 | 12 | 1955 | 383 | 18 | 4137 |
| 12 | 12 | 12_0 | COMPLETED | 24.944 | 1.0080 | 0.7018 | 17.644 | 1024 | 8 | 2631 | 367 | 21 | 2135 |
| 13 | 13 | 13_0 | COMPLETED | 24.301 | 1.0172 | 0.7420 | 18.340 | 1768 | 22 | 2927 | 77 | 10 | 4208 |
| 14 | 14 | 14_0 | COMPLETED | 23.929 | 0.9692 | 0.6905 | 16.013 | 1598 | 29 | 2592 | 396 | 23 | 3688 |
| 15 | 15 | 15_0 | COMPLETED | 24.251 | 0.9949 | 0.6952 | 16.773 | 691 | 14 | 1661 | 248 | 19 | 3523 |
| 16 | 16 | 16_0 | COMPLETED | 20.114 | 0.6694 | 0.5930 | 7.984 | 1745 | 28 | 1098 | 108 | 16 | 2467 |
| 17 | 17 | 17_0 | COMPLETED | 25.454 | 0.9170 | 0.7359 | 17.178 | 2111 | 31 | 2065 | 349 | 27 | 2251 |
| 18 | 18 | 18_0 | COMPLETED | 25.549 | 0.9484 | 0.7094 | 17.190 | 621 | 26 | 1221 | 51 | 26 | 3907 |
| 19 | 19 | 19_0 | COMPLETED | 24.824 | 0.9231 | 0.8109 | 18.583 | 1442 | 9 | 1297 | 306 | 9 | 3972 |
| 20 | 20 | 20_0 | COMPLETED | 24.913 | 0.8165 | 0.7027 | 14.294 | 810 | 33 | 1776 | 258 | 6 | 4599 |
| 21 | 21 | 21_0 | COMPLETED | 24.870 | 0.9743 | 0.7497 | 18.165 | 1308 | 32 | 2558 | 146 | 12 | 3111 |
| 22 | 22 | 22_0 | COMPLETED | 25.625 | 1.0253 | 0.7546 | 19.824 | 1333 | 20 | 1878 | 212 | 24 | 3386 |
| 23 | 23 | 23_0 | COMPLETED | 25.641 | 1.0022 | 0.7049 | 18.115 | 863 | 34 | 1535 | 104 | 25 | 3160 |
| 24 | 24 | 24_0 | COMPLETED | 24.788 | 0.9533 | 0.7081 | 16.732 | 675 | 26 | 1604 | 66 | 21 | 2720 |
| 25 | 25 | 25_0 | COMPLETED | 24.213 | 1.0209 | 0.7637 | 18.876 | 1420 | 12 | 2015 | 277 | 7 | 3621 |
| 26 | 26 | 26_0 | COMPLETED | 24.022 | 0.9163 | 0.7127 | 15.688 | 1972 | 24 | 2137 | 218 | 16 | 2822 |
| 27 | 27 | 27_0 | COMPLETED | 16.955 | 0.6163 | 0.4488 | 4.690 | 2349 | 15 | 1315 | 35 | 18 | 2282 |
| 28 | 28 | 28_0 | COMPLETED | 24.370 | 0.9586 | 0.7501 | 17.522 | 2211 | 10 | 2465 | 156 | 13 | 2585 |
| 29 | 29 | 29_0 | COMPLETED | 24.711 | 1.0388 | 0.7582 | 19.463 | 1632 | 20 | 2233 | 328 | 14 | 3025 |
| 30 | 30 | 30_0 | COMPLETED | 23.312 | 0.9011 | 0.6478 | 13.608 | 1881 | 25 | 1449 | 360 | 10 | 3843 |
| 31 | 31 | 31_0 | COMPLETED | 23.852 | 0.9480 | 0.7195 | 16.270 | 953 | 13 | 1167 | 177 | 27 | 3463 |
| 32 | 32 | 32_0 | COMPLETED | 25.429 | 1.1523 | 0.7840 | 22.973 | 1474 | 14 | 2324 | 250 | 19 | 3910 |
| 33 | 33 | 33_0 | COMPLETED | 24.912 | 1.1458 | 0.7811 | 22.298 | 1515 | 16 | 2676 | 269 | 6 | 3552 |
| 34 | 34 | 34_0 | COMPLETED | 25.132 | 1.1228 | 0.7614 | 21.484 | 1165 | 23 | 2063 | 241 | 32 | 2863 |
| 35 | 35 | 35_0 | COMPLETED | 25.680 | 1.0820 | 0.7335 | 20.381 | 1913 | 13 | 2671 | 221 | 22 | 5000 |
| 36 | 36 | 36_0 | COMPLETED | 24.773 | 1.0860 | 0.7178 | 19.312 | 1421 | 8 | 2021 | 279 | 25 | 3002 |
| 37 | 37 | 37_0 | COMPLETED | 24.990 | 1.0630 | 0.7175 | 19.060 | 1840 | 5 | 2401 | 360 | 10 | 3825 |
| 38 | 38 | 38_0 | COMPLETED | 25.533 | 1.0700 | 0.7514 | 20.529 | 1476 | 13 | 2036 | 242 | 25 | 5000 |
| 39 | 39 | 39_0 | COMPLETED | 24.830 | 1.0592 | 0.7310 | 19.226 | 1096 | 20 | 2295 | 246 | 17 | 2231 |
| 40 | 40 | 40_0 | COMPLETED | 25.093 | 1.1027 | 0.7718 | 21.355 | 2126 | 21 | 2903 | 266 | 9 | 5000 |
| 41 | 41 | 41_0 | COMPLETED | 24.613 | 1.1195 | 0.7264 | 20.017 | 1600 | 18 | 2444 | 187 | 32 | 4233 |
| 42 | 42 | 42_0 | COMPLETED | 24.369 | 1.0772 | 0.6713 | 17.623 | 1184 | 29 | 1946 | 191 | 35 | 4204 |
| 43 | 43 | 43_0 | COMPLETED | 23.966 | 1.1008 | 0.6978 | 18.409 | 1210 | 18 | 1933 | 325 | 21 | 2000 |
| 44 | 44 | 44_0 | COMPLETED | 23.437 | 1.0966 | 0.7084 | 18.206 | 1220 | 15 | 2546 | 198 | 11 | 2783 |
| 45 | 45 | 45_0 | COMPLETED | 24.425 | 1.1057 | 0.7500 | 20.256 | 1570 | 5 | 2329 | 210 | 19 | 4466 |
| 46 | 46 | 46_0 | COMPLETED | 24.617 | 1.0551 | 0.6368 | 16.540 | 925 | 22 | 2808 | 268 | 5 | 2000 |
| 47 | 47 | 47_0 | COMPLETED | 23.938 | 1.1056 | 0.7443 | 19.700 | 1631 | 22 | 2902 | 281 | 5 | 3450 |
| 48 | 48 | 48_0 | COMPLETED | 24.050 | 0.9692 | 0.6917 | 16.123 | 1720 | 5 | 2707 | 260 | 10 | 4488 |
| 49 | 49 | 49_0 | COMPLETED | 23.916 | 1.1053 | 0.7251 | 19.168 | 1451 | 9 | 2149 | 256 | 27 | 3416 |
| 50 | 50 | 50_0 | COMPLETED | 24.857 | 1.1243 | 0.7888 | 22.046 | 2358 | 19 | 2981 | 298 | 13 | 3658 |
| 51 | 51 | 51_0 | COMPLETED | 24.510 | 1.0840 | 0.7587 | 20.158 | 1490 | 19 | 2611 | 270 | 5 | 4888 |
| 52 | 52 | 52_0 | COMPLETED | 23.600 | 1.1120 | 0.7749 | 20.336 | 1790 | 20 | 2696 | 227 | 5 | 5000 |
| 53 | 53 | 53_0 | COMPLETED | 23.336 | 1.0387 | 0.7318 | 17.739 | 1120 | 18 | 2238 | 273 | 31 | 3881 |
| 54 | 54 | 54_0 | COMPLETED | 24.658 | 1.1007 | 0.7607 | 20.647 | 1509 | 18 | 2552 | 254 | 17 | 4265 |
| 55 | 55 | 55_0 | COMPLETED | 24.566 | 1.0401 | 0.7475 | 19.101 | 1317 | 18 | 2336 | 262 | 23 | 4092 |
| 56 | 56 | 56_0 | COMPLETED | 24.468 | 1.1097 | 0.7705 | 20.922 | 1671 | 16 | 2613 | 271 | 17 | 3640 |
| 57 | 57 | 57_0 | COMPLETED | 24.306 | 1.0880 | 0.7490 | 19.808 | 1596 | 19 | 2668 | 237 | 12 | 4999 |
| 58 | 58 | 58_0 | COMPLETED | 23.984 | 1.0320 | 0.7191 | 17.798 | 1428 | 15 | 2488 | 292 | 13 | 4281 |
| 59 | 59 | 59_0 | COMPLETED | 24.109 | 1.0785 | 0.7565 | 19.671 | 1513 | 15 | 2388 | 198 | 18 | 4580 |
| 60 | 60 | 60_0 | COMPLETED | 24.934 | 1.1210 | 0.7600 | 21.242 | 2134 | 20 | 2989 | 238 | 18 | 4014 |
| 61 | 61 | 61_0 | COMPLETED | 24.086 | 1.0857 | 0.7200 | 18.827 | 1568 | 20 | 2755 | 238 | 17 | 4749 |
| 62 | 62 | 62_0 | COMPLETED | 25.671 | 1.1022 | 0.7315 | 20.698 | 1647 | 14 | 2413 | 293 | 25 | 4326 |
| 63 | 63 | 63_0 | COMPLETED | 24.424 | 1.0295 | 0.7029 | 17.674 | 1403 | 18 | 2463 | 256 | 25 | 3426 |
| 64 | 64 | 64_0 | COMPLETED | 24.306 | 1.0286 | 0.7129 | 17.823 | 1303 | 17 | 2400 | 245 | 17 | 4827 |
| 65 | 65 | 65_0 | COMPLETED | 24.573 | 1.0604 | 0.6839 | 17.820 | 1677 | 19 | 2765 | 267 | 16 | 4181 |
| 66 | 66 | 66_0 | COMPLETED | 25.938 | 1.0426 | 0.7023 | 21.257 | 1553 | 13 | 2396 | 273 | 16 | 3785 |
| 67 | 67 | 67_0 | COMPLETED | 25.413 | 1.0169 | 0.7461 | 19.282 | 2579 | 21 | 3000 | 228 | 13 | 5000 |
| 68 | 68 | 33_0 | COMPLETED | 25.744 | 1.1467 | 0.7887 | 23.285 | 1515 | 16 | 2676 | 269 | 6 | 3552 |
| 69 | 69 | 34_0 | COMPLETED | 25.791 | 1.1622 | 0.7916 | 23.729 | 1165 | 23 | 2063 | 241 | 32 | 2863 |
| 70 | 70 | 35_0 | COMPLETED | 25.335 | 1.0285 | 0.7429 | 19.358 | 1913 | 13 | 2671 | 221 | 22 | 5000 |
| 71 | 71 | 36_0 | COMPLETED | 25.441 | 1.1046 | 0.7649 | 21.494 | 1421 | 8 | 2021 | 279 | 25 | 3002 |
| 72 | 72 | 37_0 | COMPLETED | 25.850 | 1.0397 | 0.7647 | 20.553 | 1840 | 5 | 2401 | 360 | 10 | 3825 |
| 73 | 73 | 38_0 | COMPLETED | 25.261 | 1.0881 | 0.7599 | 20.888 | 1476 | 13 | 2036 | 242 | 25 | 5000 |
| 74 | 74 | 39_0 | COMPLETED | 25.638 | 1.1548 | 0.7812 | 23.128 | 1096 | 20 | 2295 | 246 | 17 | 2231 |
| 75 | 75 | 46_0 | COMPLETED | 25.874 | 1.1494 | 0.7752 | 23.055 | 925 | 22 | 2808 | 268 | 5 | 2000 |
| 76 | 76 | 76_0 | RUNNING | NaN | NaN | NaN | NaN | 1154 | 29 | 1883 | 262 | 31 | 2649 |
| 77 | 77 | 77_0 | RUNNING | NaN | NaN | NaN | NaN | 1146 | 25 | 1924 | 256 | 32 | 2734 |
| 78 | 78 | 78_0 | RUNNING | NaN | NaN | NaN | NaN | 1148 | 31 | 1972 | 242 | 33 | 2633 |
| 79 | 79 | 79_0 | RUNNING | NaN | NaN | NaN | NaN | 1181 | 31 | 1958 | 263 | 32 | 2777 |
| 80 | 80 | 80_0 | RUNNING | NaN | NaN | NaN | NaN | 1165 | 23 | 1976 | 253 | 33 | 2692 |
| 81 | 81 | 81_0 | RUNNING | NaN | NaN | NaN | NaN | 1164 | 23 | 1962 | 268 | 34 | 2712 |
Insight Analyses display information to help understand the underlying experiment i.e parameter and metric relationships learned by the Ax model.Use this information to better understand your experiment space and users.
The top surfaces analysis displays three analyses in one. First, it shows parameter sensitivities, which shows the sensitivity of the metrics in the experiment to the most important parameters. Subsetting to only the most important parameters, it then shows slice plots and contour plots for each metric in the experiment, displaying the relationship between the metric and the most important parameters.
Sensitivity Analysis for Pmax
Understand how each parameter affects Pmax according to a second-order sensitivity analysis.
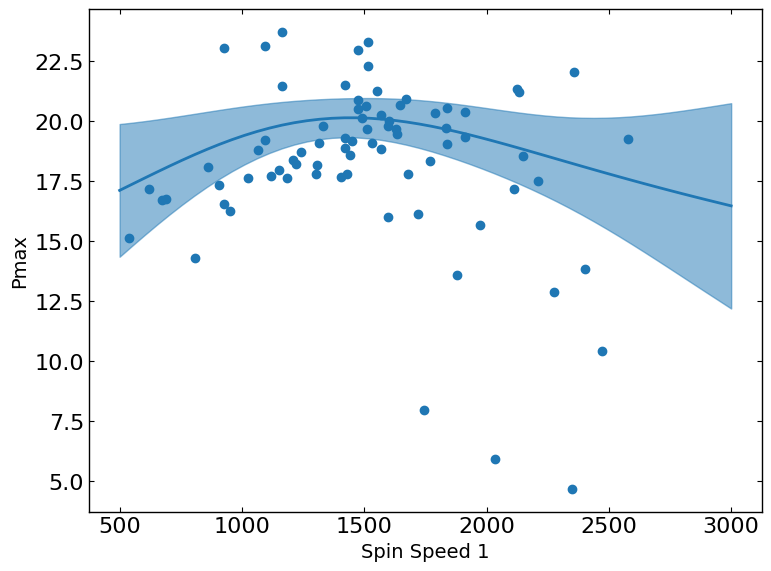
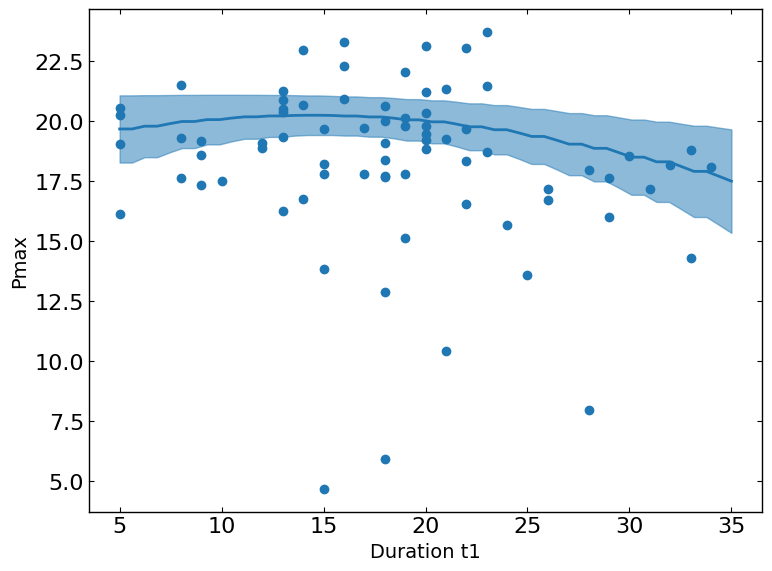
These plots show the relationship between a metric and a parameter. They show the predicted values of the metric on the y-axis as a function of the parameter on the x-axis while keeping all other parameters fixed at their status_quo value (or mean value if status_quo is unavailable).
Pmax vs. Spin_Speed_2
The slice plot provides a one-dimensional view of predicted outcomes for Pmax as a function of a single parameter, while keeping all other parameters fixed at their status_quo value (or mean value if status_quo is unavailable). This visualization helps in understanding the sensitivity and impact of changes in the selected parameter on the predicted metric outcomes.
Pmax vs. Spin_Speed_1
The slice plot provides a one-dimensional view of predicted outcomes for Pmax as a function of a single parameter, while keeping all other parameters fixed at their status_quo value (or mean value if status_quo is unavailable). This visualization helps in understanding the sensitivity and impact of changes in the selected parameter on the predicted metric outcomes.
These plots show the relationship between a metric and two parameters. They show the predicted values of the metric (indicated by color) as a function of the two parameters on the x- and y-axes while keeping all other parameters fixed at their status_quo value (or mean value if status_quo is unavailable).
Pmax vs. Spin_Speed_1, Spin_Speed_2
The contour plot visualizes the predicted outcomes for Pmax across a two-dimensional parameter space, with other parameters held fixed at their status_quo value (or mean value if status_quo is unavailable). This plot helps in identifying regions of optimal performance and understanding how changes in the selected parameters influence the predicted outcomes. Contour lines represent levels of constant predicted values, providing insights into the gradient and potential optima within the parameter space.
Diagnostic Analyses provide information about the optimization process and the quality of the model fit. You can use this information to understand if the experimental design should be adjusted to improve optimization quality.
Cross-validation plots display the model fit for each metric in the experiment. The model is trained on a subset of the data and then predicts the outcome for the remaining subset. The plots show the predicted outcome for the validation set on the y-axis against its actual value on the x-axis. Points that align closely with the dotted diagonal line indicate a strong model fit, signifying accurate predictions. Additionally, the plots include confidence intervals that provide insight into the noise in observations and the uncertainty in model predictions.
NOTE: A horizontal, flat line of predictions indicates that the model has not picked up on sufficient signal in the data, and instead is just predicting the mean.
Cross Validation for Pmax
The cross-validation plot displays the model fit for each metric in the experiment. It employs a leave-one-out approach, where the model is trained on all data except one sample, which is used for validation. The plot shows the predicted outcome for the validation set on the y-axis against its actual value on the x-axis. Points that align closely with the dotted diagonal line indicate a strong model fit, signifying accurate predictions. Additionally, the plot includes 95% confidence intervals that provide insight into the noise in observations and the uncertainty in model predictions. A horizontal, flat line of predictions indicates that the model has not picked up on sufficient signal in the data, and instead is just predicting the mean.
[11]:
from ax.utils.notebook.plotting import render# for plotting in notebook
from ax.plot.slice import plot_slice
from ax.plot.scatter import interact_fitted
from ax.adapter.cross_validation import cross_validate
from ax.plot.diagnostic import interact_cross_validation
from sklearn.metrics import r2_score
model = optimizer.ax_client._generation_strategy.adapter # get the model from the ax client
cv_results = cross_validate(model)
# calculate the r2
observed, predicted = [], []
for i in range(len(cv_results)):
observed.append(cv_results[i].observed.data.means[0])
predicted.append(cv_results[i].predicted.means[0])
r2 = r2_score(observed, predicted)
print(f'R2 = {r2}')
render(interact_cross_validation(cv_results))
obj_type = 'Pmax' # objective type
p_idx = 0
for p_idx in range(len(optimizer.params)):
x = ax.plot.slice.plot_slice(model=model,param_name= optimizer.params[p_idx].name, metric_name= obj_type).data['data'][1]['x']
y = ax.plot.slice.plot_slice(model=model,param_name=optimizer.params[p_idx].name, metric_name= obj_type).data['data'][1]['y']
x1 = ax.plot.slice.plot_slice(model=model,param_name= optimizer.params[p_idx].name, metric_name= obj_type).data['data'][0]['x']
y1 = ax.plot.slice.plot_slice(model=model,param_name= optimizer.params[p_idx].name, metric_name= obj_type).data['data'][0]['y']
# cut x1 and y1 to the same length as x and y
x_low = x1[:len(x)]
y_low = y1[:len(y)]
x_high = x1[len(x):]
y_high = y1[len(y):][::-1]
plt.figure(figsize=(8,6))
plt.plot(x,y)
# shade the area between the two curves
plt.fill_between(x, y_low, y_high, color='C0', alpha=0.5)
# plt.fill_between(x, y, y_high, color='gray', alpha=0.5)
plt.plot(data[optimizer.params[p_idx].name],data[obj_type],'C0o')
plt.xlabel(optimizer.params[p_idx].display_name)
plt.ylabel(obj_type)
plt.tight_layout()
plt.show()
R2 = 0.5015176370786073
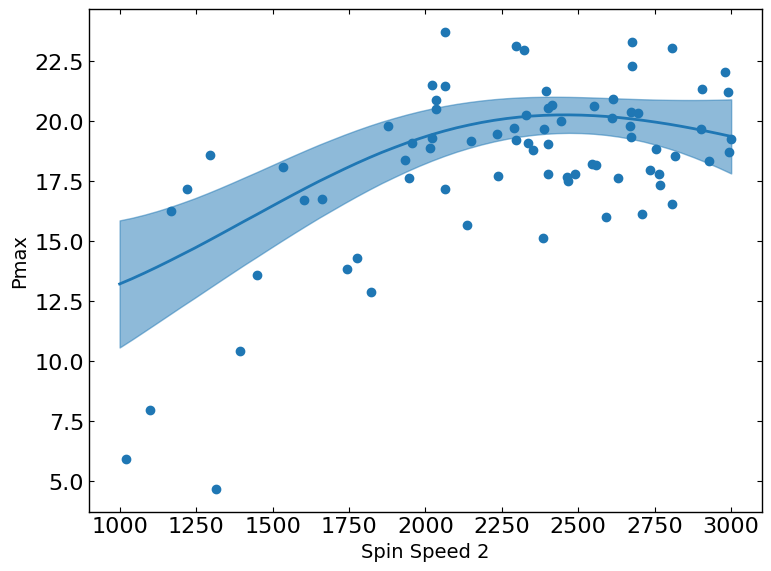
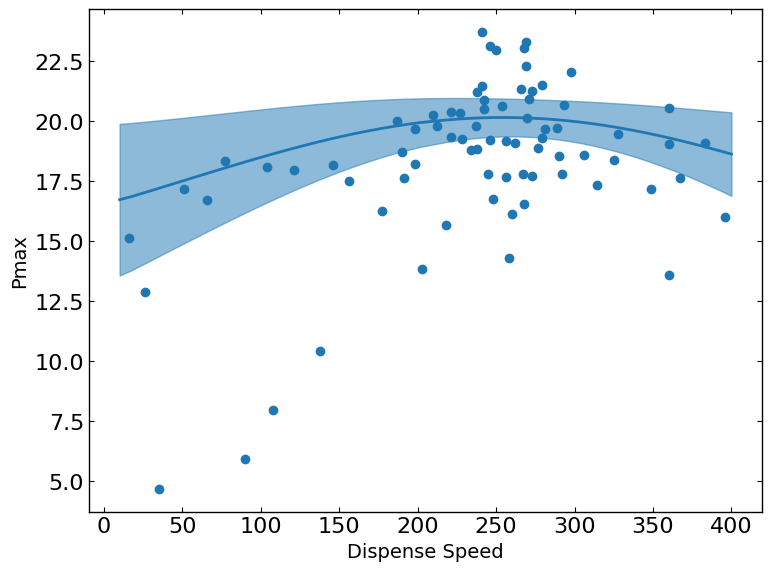
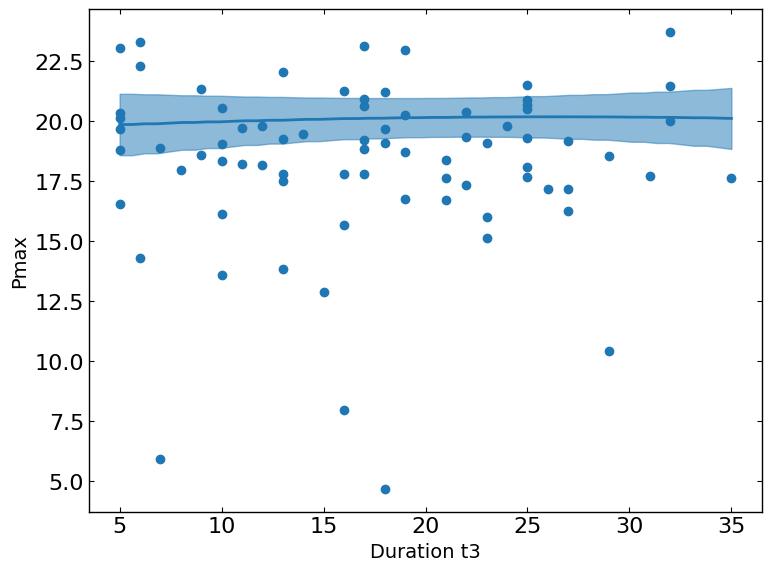
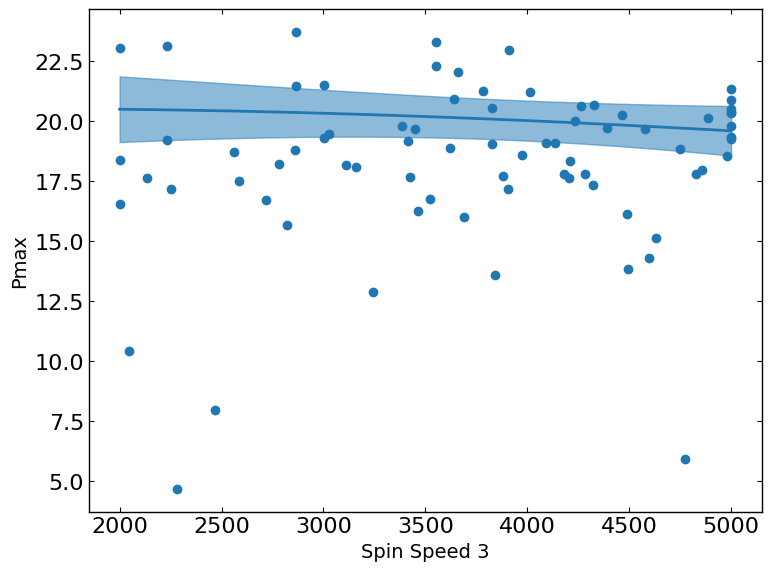
[12]:
# Plot the density of the exploration of the parameters
# this gives a nice visualization of where the optimizer focused its exploration and may show some correlation between the parameters
plot_dens = True
if plot_dens:
from optimpv.posterior.exploration_density import *
params_orig_dict, best_parameters = {}, {}
for p in optimizer.params:
best_parameters[p.name] = p.value
fig_dens, ax_dens = plot_density_exploration(params, optimizer = optimizer, best_parameters = best_parameters, optimizer_type = 'ax')